Episode 9 (bonus)
If you enjoyed this episode or at least learned something new, please consider supporting the podcast by buying me a coffee.
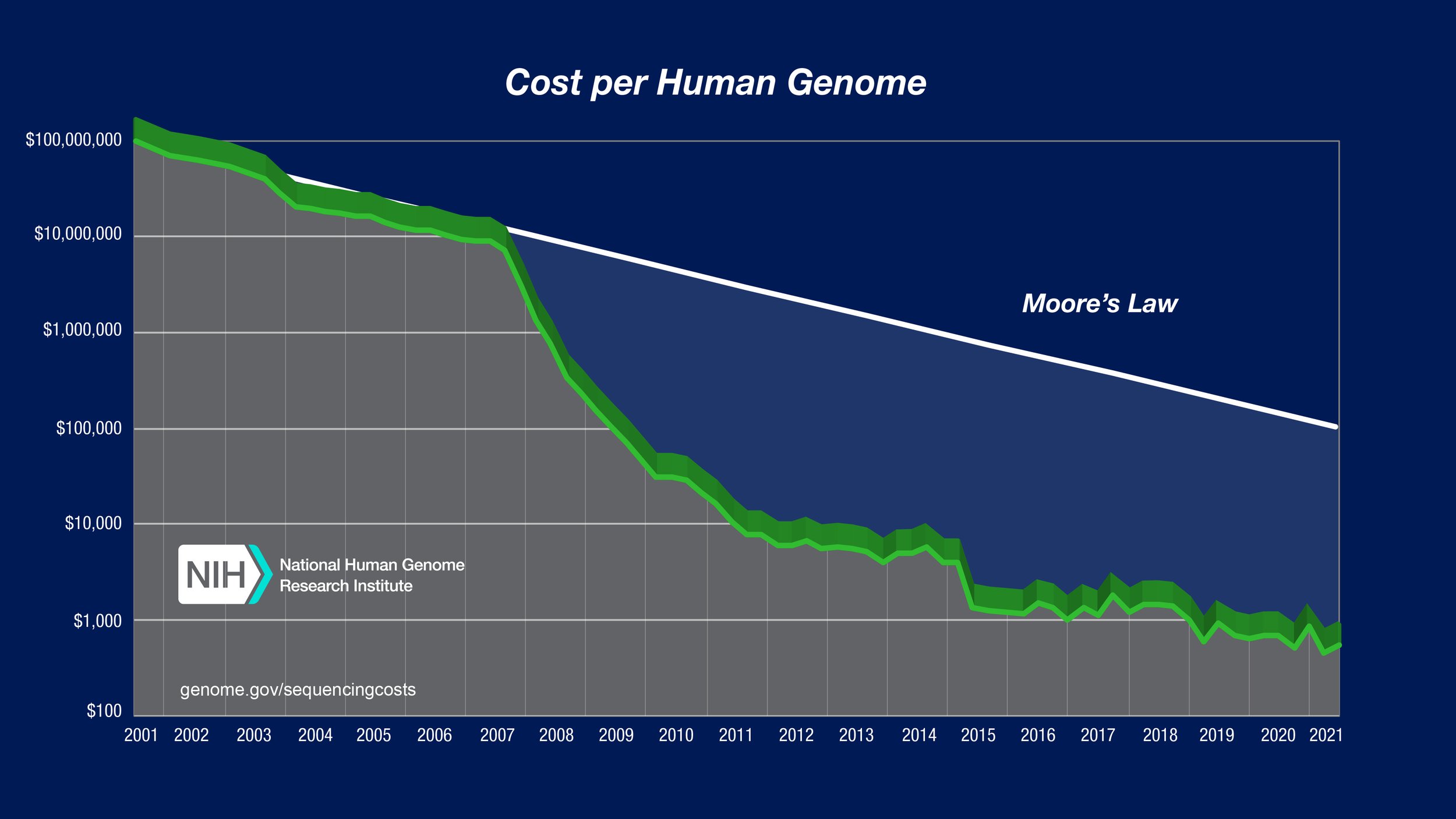
Cost per genome data - 2021, visit https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost for further information.
Massively Parallel Sequencing has revolutionized laboratories' ability to harness more and more information from our genetics. This has led to massive improvements in medical diagnostics, public health (think SARs-CoV-2 variant identification), ancestry determinations, and now forensic science has begun adapting this technology to criminal investigations.
In this episode I speak with Professor Megan Foley, Teresa Snyder-Leiby, and Sarah Copeland about the challenges laboratories are facing as they adopt this type of DNA testing and tools that are available not only to labs, but also to law enforcement, prosecutors, and defense - including the Innocence Project.
This episode is in partnership with the Association of Forensic DNA Analysts and Administrators (AFDAA). Visit afdaa.org to learn more about this professional organization.
Are you part of a not for profit organization and would like to have a featured episode promoting your organization or upcoming events, please email hello@secretsfromthecrimelab.
To learn more about the guests, see their bios at https://practicalforensics.science/podcast. Simply scroll down to the post for episode 9.
Megan Foley is a visiting assistant professor in Forensic Molecular Biology at the George Washington University. She has her bachelor of arts in biology from The College of St. Benedict, her masters of science in forensic science from Arcadia University, and is currently working on her PhD in forensic science from Oklahoma State University. Before teaching for GW, she worked as a forensic biologist at NMS Labs performing casework and method validation, and was the DNA Training Specialist and Lead DNA Researcher for The Center for Forensic Science Research and Education. Her current research focuses on evaluating new products in order to develop recommendations and strategies for laboratories for implementation. Especially focusing on next generation sequencing platforms and analysis software in order to develop analytical parameter recommendations for analysis and probabilistic genotyping.
Sarah Copeland is Senior Biologist SoftGenetics (3+ years) involved in software development, testing and support for forensic fragment CE data analysis and MPS data analysis.
Education: UC Davis, Forensic Science Program Master of Science in Forensic Science , thesis research: Mitochondria DNA Analysis of Highly Degraded Bone Samples Using Next Generation Sequencing (employed Mitochondrial DNA (mtDNA) as well as nuclear DNA (nDNA) analysis, Sanger Sequencing and capillary electrophoresis, Next Generation Sequencing (NGS) including Roche 454 and Illumina MiSeq, Library preparation for both amplicon and whole genome probe capture NGS,DNA extraction from bone, buccal swabs, hair, blood, and tissue samples. She was previsouly employed by Mitotyping Technologies DNA Analyst (4 years) before joining Softgenetics.
Teresa Snyder- Leiby (one of those older guys with about ½ her career in molecular bio, the last half in forensic DNA analyses). Her degrees (BS, MS, PhD) are from Juniata College, North Dakota State Univ. and Penn State. She was a technician at Penn State way back when RFLP was the new DNA fingerprinting method (mid-1980s)….. in plant physiology program she worked on RFLP analysis of Rose varieties for the Horticulture department – case never came to court as the defendant removed their variety from the market when notified DNA analysis results would be presented. Her thesis research was with isolation and sequencing genes involved in cacao disease resistance. After graduate school she was a biology professor at Marist College and State University of New York (general biology, environmental microbiology, electron microscopy and biotechnology including human forensic microsatellite analysis and kinship statistics). She joined SoftGenetics in 2008, and as a technical forensic specialist; provided technical assistance to forensic laboratories with genotyping and relationship testing statistics. Moved on to product manager for forensic genotyping software and more recently development and validation of MPS forensic data analysis software and probabilistic genotyping software.
Resources
https://softgenetics.com/
Review article about MPS and mitochrondrial MPS https://wires.onlinelibrary.wiley.com/doi/epdf/10.1002/wfs2.1431
Norah Rudin and Keith Inman book mentioned in the episode https://www.amazon.com/Introduction-Forensic-DNA-Analysis/dp/0849302331
Also visit ISOGG for more information about mitochondrial DNA, haplogroups, etc https://isogg.org/wiki/Wiki_Welcome_Page
Join our discussion and let us know your thoughts.
E: hello@secretsfromthecrimelab.com
OR
Join our Facebook Page! https://www.facebook.com/SecretsfromtheCrimeLab
Instagram: secretscrimelab
Comments? Feedback? Suggestions? Or if you would like to be a guest?
E: hello@secretsfromthecrimelab.com
Comments? Feedback? Suggestions? Or if you would like to be a guest?
Get in touch on the Contact Page
Transcript (presented without fixing the translation program’s abominable punctuation)
Ep 9 (S1 Bonus)
[00:00:00] Angela: Greetings all and welcome to another episode of Secrets from the Crime Lab an educational podcast, where we inform an encourage discussion on forensic science and related scientific fields and topics. I'm your host, Angela Swarts. I have three guests with me today, which I will introduce to everyone shortly, but first I wanted to do a short little announcement for everyone. I did complete season one. And so this is one of two bonus episodes that we're going to have. We had some scheduling difficulties related to people like myself, having the flu and other people having COVID. So things have been a little bit delayed, but so we're gonna have two bonus episodes for season one that I'll have out for you.
I'm scheduling guests for season two at the moment. So that will be sometime this fall when those episodes will be available. In the meantime, I wanted to thank everyone for tuning in and listening. We've already crossed the threshold of a thousand downloads and I really haven't done any marketing.
[00:00:59] Angela: So that's really amazing for all of the support. And I couldn't have done that without everyone sharing this around with their friends. So I really appreciate that. I have had some requests to do video content to go along with the podcast. At the moment this podcast is completely self-funded. So if video content is something that you would like to see in the future, I would appreciate your support to get there. If you would consider making a donation, to help cover production costs. if you're listening on a podcast app, there's a little link at the end of the show notes that says support the show. That'll take you to some place where you can make a donation in whatever quantity you want. Currently I'm using Buy Me a Coffee, so you can literally buy me a coffee, but I might change that to buy me a taco since I'm moving back to Texas. And if you're listening on the web player, there's a little heart icon next to the social media links, and you can just click on the heart and it will take you to the same place.
Okay. And so with the announcements out of the way, let's move on to our topic for today. So I'm doing this in partnership with the Association of Forensic DNA Analysts and Administrators. Their annual summer meeting is taking place in Houston at Hotel, July 28th and 29th, I believe. I'll put a link in the show notes, 'cause I may have those dates incorrect for anyone who's interested in attending that. So the day before the meeting, there is a free workshop on the analysis and validation of forensic MPS data, and MPS is massively parallel sequencing. And that is what we're going to be talking about today. That workshop is hosted by Soft Genetics and it's Wednesday, July 27th from one to 5:00 PM. And so we're gonna just talk a little bit about what that workshop is going to be about. It is gonna be a pretty technical workshop, but it is open to anyone who wants to go. So whether or not you're a scientist, if you're a prosecutor defense or in the Houston area, and really interested in that sort of thing, you can certainly RSVP for it, but it is gonna be technical. So you are forewarned on that.
So massively parallel sequencing is something that generates a lot of data. And that's something that we're gonna talk about later on in the podcast. Soft Genetics is a company that I'm gonna allow Teresa to talk bit about what So software solutions for data analysis. And I also have her colleague, Sarah, with us today as well. and then we have Megan who's a visiting professor at George Washington University at the moment. And I'm going to hand it over to Megan to start with and let her introduce herself to everyone.
[00:03:49] Megan Foley: All right. So hello. Thank you for having me. So yeah, I can talk a little bit about how I got to where I am today. So I, have a bachelor of arts and biology from college of St. Benedict, which is a small private school in Minnesota, which is where my home is. And then I went and got my master's degree in forensic science at Arcadia University. So at that time, after my master's, I went and started working in a private crime lab in the area doing forensic biology casework. So I did a lot of different types of casework from pretty much start to finish from opening a case to reporting. And then I also got into a lot of the validation work. So testing the new products that are coming out in the field and getting them ready to run casework on. And so validation if you've ever done it, it's kind of on the research side of things. I love it. Not a lot of people will tell you that I do love validations. But because of that, I actually decided to go into more the research side of forensics. So I switched to a different department at the company focusing on training analysts, performing research, and then the education part. So during my time there, I decided I wanna go back and get my PhD. So I'm working on that right now through Oklahoma State University, in forensic science. And then I took a job here at George Washington University in DC as a visiting professor, teaching for their master's program in forensic science.
So a lot of the research i've done because I have bridged both worlds of casework and research. I can see it from a different perspective of how do you implement the research that's going on. There is some awesome research going on by institutions and universities, but how do we get it into the crime lab then? Because there's a lot that goes in between, about it, but then getting it ready to go to court. So I have focused a lot of my research on whether it's new products or current products testing the limits out.
And finding ways that labs can take the research I've done and use it to implement in the lab, tell them, you shouldn't consider getting this instrument or this piece of machinery or consider this when you're validating it. So a lot of it could be a new product. A lot of what I'm focusing now is on the implementation of MPS. So it's a whole new realm of data and statistics. So that's really what I have been doing right now.
[00:06:14] Angela: All right. Thank you. Thank you for coming on the podcast. I appreciate it. Sarah, you wanna go next? Introduce yourself.
[00:06:20] Sarah Copeland: So let's see, I got my bachelor's in, physiology actually at San Francisco State University. I took a little break in between, and before I got my master's and I did that at, university of California Davis, and that was in forensic science, focusing on DNA analysis and I did my thesis work was NGS analysis of degraded bone samples. After that I started working at Mitotyping Technologies for a few years as a forensic DNA analyst. And so Mitotyping specifically does specifically does mitochondrial DNA work. So that's mainly you know, hair, shafts, things like that. And so I worked on that from extraction, through the whole process to Writing the reports and testifying in court. After that, I moved on to Soft Genetics and here I get to help people, troubleshoot I help them learn how our software and, things like that. So it's, it's a lot of, training and education for people that are, they're using the software in their day to day work.
Yeah. I do a lot of the training so that they can use it day to day. If they get difficult samples that they're unsure of what to make of them. I work with them to, to figure that out. So it's, it's always interesting.
[00:07:50] Angela: So you're the phone, a friend.
[00:07:52] Sarah Copeland: Yes.
[00:07:53] Angela: you're the phone a friend for the, is that TV show even still on anymore? I don't even remember what it was called with the phone a friend thing…
[00:08:00] Megan Foley: wants to who millionaire
[00:08:01] Angela: Oh yeah. Who [wants to] be a millionaire, right? Yeah. So I think all of us would like to be a millionaire. All right. Teresa?
[00:08:07] Teresa Snyder-Leiby: Yes. Thanks so much for, uh having us on your podcast. And I started my academic career different area, ecology and environmental science for my bachelor's degree.and then worked as technician, various positions. By the time I got to Penn state, switched into molecular biology of disease resistance. and so that. involved Developing PCR, primers. we did a lot of sanger sequencing and a lot of basically gene discovery in, in disease resistance in agriculture. and then for about 15 years, I was a biology and, biotechnology professor up in New York state. We moved back to Pennsylvania and, switched gears and have been working for Soft Genetics for about the last 15 years where we develop forensic, DNA analysis software for any of the different platforms. There's another side of the aisle where we also are very active with basic research software. So any type of MPS research, but that's separate from the forensic 'cause the forensic has specific needs. In our software development, as Sarah was saying, we do a lot of training support, assistance, and interaction with the labs that are using our software. So helping them to get the most out of the software and also getting suggestions for them, of what functions would be helpful to include in new versions. So it's, a lot of, it's a lot of teaching, but not in that university lecture hall lab type of setting. It's, much more one on one or one to small group and in very specific areas, Depending on the where the laboratory has, you know, what their research or what their lab and data analysis requirements are.
[00:09:55] Angela: So it's, professional on the job type trainings for specific applications, whether it is in a research laboratory or in a, production forensic laboratory?
[00:10:05] Teresa Snyder-Leiby: Yes, absolutely. Yeah. Unlike the freshman classes, no one asks me if they have to learn this or will it be on the test? You that's 'cause it's very refreshing to interact with professionals who are very motivated to become skilled and understand what they're working with.
[00:10:24] Angela: All right, in everyone's introductions, we threw out a lot of jargon. Like DNA nerd jargon that not everyone that listens is going to know what it is. So we've said MPS we've said NGS. Does anyone wanna explain what those are?
[00:10:44] Teresa Snyder-Leiby: Basically NGS and MPS are two different abbreviations for the same thing. It was originally I think, called NGS, which stood for next generation sequencing, but it's now been around for 15 or more years. So it's not really a good name for it. MPS is much more descriptive. It stands for massively parallel sequencing. And so that describes what's going on is that you've got many, many, many, many, many different sequencing events going on all at the same time. And so you're gathering this depth of sequence information that prior to this technology being developed was not possible to have that depth of information. And actually historically the development of, of MPS is why the human genome project that probably people remember back when we were scientific community was undertaking sequencing the human genome. it actually finished ahead of schedule because they originally thought they would have to use Sanger sequencing for the whole thing MPS was developed. And so that kicked everything into high gear.
[00:11:57] Angela: And so we're talking about two different types of sequencing. So we say the old sequencing was Sanger type sequencing. Sanger was a person's name that. That developed the technique. And that's where we're just looking at a single stretch on one piece of DNA, basically to start with. And we're looking at that individual sequence of that small section. Back before we even had automated, we were doing manual sequencing where we were even using radioactive materials to label it and stuff. So we would get short stretches 300 to 600 bases. But when we're talking about massively parallel sequencing, we were saying it's massive. And in parallel, because we're sequencing all of these different stretches all at the same time. Just in multiple samples at the same time. Yes,the throwing all of your sample into one bin with , biochemical barcodes on them. It's definitely a paradigm shift for those of us that work in a forensic laboratory where we're like, no, everything has to be kept separate. You can't do that. And I know some of this is gonna be, we're trying, even though we're trying to keep this as a high level discussion, I know that some of the technical side of things, it's hard for us not to get a little bit technical in some of this, some of the conversation. But they, the MPS was a massive leap forward technologically, for anybody who was doing any kind of DNA work, right? Any kind of research like you used to do agriculture research. I used to do cancer research on specific, tumor genes before I went into forensics. And that's one of the things. Those of us who are interested in forensics or in the forensic field. Some sometimes forget 'cause we get very focused on forensics, but all of these new technologies mostly developed in research and clinical side and then forensics goes and looks at and goes, Hey, this is really neat technology. We can do a lot with this. Let's just tweak the sensitivity. ‘Cause we have degraded samples and low quantity samples. How do we make this technology that you guys have figured out over here on the clinical side? Now let's take, pull that into forensics. Now that you've worked all the kinks out of it and make a little bit more specialized for us. And so that's one of the things that we talk about. There's a there's aspects of this technology that are really helpful for, Sarah mentioned that she worked with, bone samples. Quite a bit and hair shafts and that's specifically, usually mitochondrial DNA. So, Sarah, could you tell us a little bit about why we wanna use mitochondrial DNA when we're looking at bones and hair shaft?
Sure. Bones, hair, shafts. They they, don't have a whole lot of, nuclear DNA. That's the stuff that comes from your mother and your father and, creates who you are. There's a lot of DNA present in your mitochondria and that, there's a lot of mitochondrial DNA, a lot of mitochondria in general in, bones. And so it's a lot easier to get that information, from those samples rather than the nuclear DNA, which isn't really present.
[00:15:09] Angela: the mitochondria are present in like hundreds or thousands of copies in a cell. Whereas your nuclear DNA, you've only got one set of your nuclear DNA in each one of your cells. But when we're talking about like anthropological samples, like bones that have been dug up, the nuclear DNA is all degraded and gone. So the mitochondrial is the only thing we'd be able to get DNA from usually.
[00:15:32] Sarah Copeland: Yeah, exactly.
[00:15:34] Angela: so we got the mitochondrial applications and we can use MPS to test the mitochondrial DNA as well. And we can get a lot more information out of that than we could. using the older techniques. And I think, Teresa, I think you shared a paper. it was a review article where someone had gotten, a mitochondrial DNA from like a one millimeter of a hair shaft?
[00:15:58] Teresa Snyder-Leiby: Yes. Yeah. it was amazing. and Actually that's one of the presentations that will be at the workshop is that, routinely they're able to get complete Mito, haplo grouping from one to five millimeter pieces of hair. And so one of the slides shows like One millimeters, basically a pencil point, and five millimeters is the eraser. not, as Sarah said, you would not be getting nuclear DNA out of these. There's no root. there's no nuclear DNA. They tested recent samples. think samples that were five or six years old and some that were 40 to 50 years old. So they were still able to get from the 40 year old samples, which would be, in, in line with the, what do you call it? Cold cases? Some of the cold cases. So not ancient DNA, but a cold case that, body that's been found after 30 or 40 years, to be able to get that information. And so the Mito is going to give them information about the maternal lineage of that person. and if they can sequence the whole mitogenome so about 16,000 or so well, a little more base pairs from the whole genome, they can even separate, in,I think it was 40% of the cases they can separate the mother from the child. Because of, mutations that occur. And so for the most part, the mother child would have the same, Mito sequence but when they do the entire sequence, not just the control region, they're more likely to find heteroplasmies or slight differences between the mother and child. And so you can actually, in many cases, even differentiate between a very close yeah, you don't get any closer than mother, child for mito transfer. That's pretty amazing. Yeah. So that's the sort of thing that I think two centimeters is more commonly used for Mito extraction. But, they went through the very small pieces and that's one of the presentations I was amazed. That's actually impressive. And I think you're right about the two centimeters. I think that's what i've, sent off to laboratories that do that sort of work. So it's got a lot of applications for the missing persons and unidentified human remains, disaster victim identification. So if we have mass disaster is certainly gonna be helpful, 'cause I mean, you probably wouldn't be using like one to five millimeter piece of something when you've got the, those kind of things. You're not necessarily, it's more about matching up the remains the right person, but it's still amazing that you can do that. 'Cause normally with the traditional Sanger type sequencing where you were only able to look at the control region, you wouldn't be able to differentiate between the mother and the child, you need the whole genome to do that. And you can only do that with MPS. Yeah. Sanger sequencing the whole Mito genome would be way, way cumbersome. And you only get, as you were saying, one copy. So you can miss hetorplasmy and minor differences with Sanger that MPS is able to pick up.
[00:19:00] Angela: Forensic laboratories, when we look at things mitochondrial DNA, isn't what we, that's not our go to. That's not the first thing we look at. The first thing we usually look at are our standard, what we call STRs or short tandem repeats. And I know Megan, you said that a lot of your research is you're focusing more on the big picture for all of the data. Could you talk to us a little bit about STRs and also Y-STR what are they, and what are we looking at in those and how the newer technologies like the MPS is so beneficial to forensic laboratories, once they have the money to get the equipment
[00:19:37] Teresa Snyder-Leiby: Yeah.
[00:19:37] Megan Foley: That is true. Yeah, absolutely. So STRs are parts of the DNA that as humans, we each share on these sections. Now they're not genes. We don't look at genes and forensics with STRs, but they are just short base pair or building block repeats. they might go A-T-G-C as an example and repeat over and over again. And each person might have a slightly different number of how often these repeat. so that's where the whole re the short tandem repeat comes in. There are in, at least in the us, we focus in on 20 locations of these STR regions. And by gathering all that information, we can start to develop unique DNA profiles that way. so with that is your nuclear DNA. So you do have two copies, one from your mom, one from your dad. So you at each of these locations, you're gonna see two separate numbers. alleles and alleles are just different variations of whatever that gene or STR could be. so this is what we do in typical casework. If we have enough DNA, and nuclear DNA, we can create this STR profile, and then use this data. It, it comes out in looks like a graph comes out with these large peaks and then there's a number underneath it. We use that number to tell how many repeats there are. so you could have two numbers or you could have one number. And that just, again, depends on the allele that you inherited. Sometimes it is the same between both parents. and that's called a homozygous location. Heterozygous is when you have two different, numbers of repeats. That's basic autosomal STRs. We also can use Y-STRs as testing as an additional tool. Now, in your chromosomes, you have your sex chromosomes, and for females, you have two X chromosomes, a male's gonna have an X and Y so Y-STRs are STRs on the Y chromosome. So it's a tool we can use to focus in on if there's a male contributor. and this is really beneficial when we're doing cases where there is a high amount of female and maybe a low amount sexual assault cases are very common. and STRs are super useful for that in that we can. Almost block out. It's like,if you think of the radio and there's multiple frequencies, we can block out the frequency of the female to focus in on that male component of the DNA. so it just, lets us just look at that. Now, similar to what you guys talked about with mitochondrial DNA, the Y-STR profile is passed on paternally. So you are going to have the same Y-STR profile as your father, as your biological brother, as your uncle, your, father's brother. That,it just changes how we will report on the results for that. but moving into the MPS or the sequencing realm with it right now, our technologies will give us that number. It'll say ATGC repeated 15 times. now with the sequencing aspects, since we can look at every single little nucleotide, we might find that two individuals that both shared, I'm giving as an example, the number 15. now under sequencing, we might be able to see there, there's actually a difference between that 15. I use ATGC as an example, one person might have ATGC and one person might have ATCC. during our normal methods, that looks the same. With sequencing we can actually see the difference between the two much more discriminatory.
[00:23:03] Angela: I think there was, an analogy. I think I could be incorrect, but I think it was in the book forensic DNA typing, by Nora Rudin and Keith Inman. They talked about STR as a train and everyone has an engine and a caboose, but the number of cars in between changes, right? So the length of the train is the same. Everyone's got the same length of train or the, but the number of cars in between changes. And then if we're talking about the individual sequencing, we would say that the length of that train is the same, but then we can tell that the upholstery on the inside of one of those cars is a little bit different. is that kind of,
[00:23:42] Megan Foley: Absolutely wonderful. yeah.
[00:23:45] Angela: We're talking about the upholstery, but we would also talk about that. this one has a seatbelt and this one doesn't have a seatbelt and this one's got. Tinted windows. And this one doesn't have tinted windows. I mean, that started like every little detail you could possibly think of inside of those train cars. We can start to understand and visualize with MPS.
[00:24:07] Megan Foley: Yes.
[00:24:08] Angela: that adds a layer of complication to data analysis. And we need software tools and we need researchers to tease through all of that so that we know what are we doing with all of this information? How do we convey it in a courtroom setting? How do we make it useful for investigative purposes?and I know Megan, you had mentioned that you, part of your research is you've done a survey of crime laboratories and try to see where are the labs at the moment with. understanding the technology acquiring the technology, understanding what to do with all of this information.
yeah, absolutely. A colleague,Dr. Fabio Oldoni and I, he is a professor at Arcadia university, we both have experience in the sequencing realm and thought,there is so much... ev-every lab's working on implementing it in their own way, but where is everybody at? And how can we help,as on the research side of things. So we did send actually globally a survey out on just current status. do you have an NGS? but also if you don't have a sequencing instrument, what are the hurdles, what is, what are the challenges you're dealing with trying to implement it? so a lot of what I'll talk about during the workshop are some of the results we saw on, of course the biggest hurdle is funding. unfortunately we can't, I can't do much about that. as a researcher, cause we all could use a little bit more to help. but there were some other responses that a lot of the respondents did say things like, lack of standards and recommendations and, something I'll bring up during the workshop is there's actually some out there. so we're hoping that the results of the survey just help educate people, around the US, around the world of, you're not alone in this, but also there's resources you can use. and there was a lot, you know, a lot of concern about validation. So there are some SWGDAM guidelines. So some guidelines that we often follow in the forensic field, and also a big concern is how are we going to do the statistics? right now we, as a field are implementing a statistic called probabilistic genotyping. and it's not something we can do by hand. we need the software to do it. And now that we...
[00:26:21] Angela: I've I've done it by I've done it by hand. I did it for one locus on a validation. I did it by hand. I will never repeat that.
[00:26:31] Megan Foley: I believe it
[00:26:32] Angela: A week. took me a week.
[00:26:35] Megan Foley: Yeah. Our turnaround times would go way up if we had to do that. yeah, but so now that we have all of this, we have so much more data now doing the sequencing, the statistics are just going to become more complex. so there's a lot of opportunity tobridge that gap and to get these two different, technologies, techniques to come together. So we can actually do casework on sequencing data.
[00:26:58] Angela: One of the things that you mentioned, so your survey that you did, it was global. Did you see, a difference between where's,where the United States and Canada, as compared to the UK and Europe and Australia, are we, is the US, North America, are we keeping up or are we a little bit behind? Cause I know there's so much research that seems to come out Europe with MPS.
[00:27:21] Megan Foley: Yes, Yeah. Iin terms we're pretty much we're keeping up. also the big difference is the law part of it, the legal systems are different between countries. one country might be able to take to court something they do at a university lab versus in the us, we can't, we're not gonna do that. so that I think might be the, Kind the biggest difference, but overall, globally, we, we seem the field altogether seems to be in that same place of, what are the big needs that people could use to implement it.
[00:27:50] Angela: So people have either just gotten it or are trying to figure it out, or they're trying to get funding
[00:27:56] Megan Foley: mm-hmm
[00:27:57] Angela: and planning to get it. There's not. Yeah. There's not a lot of laboratories that are doing it routinely. Certainly not. I wouldn't think government laboratories, I would think it's more likely in the private labs that would be able to get the funding for these. Maybe? No? Teresa's waving at me.
[00:28:14] Teresa Snyder-Leiby: I,I did just wanna say there is a publication out of the FBI where they validated, the control region of mito with MPS for casework.
[00:28:23] Megan Foley: Wonderful.
[00:28:24] Teresa Snyder-Leiby: Yeah, it's relatively new. that's why I was waving at you. Like, wait. I know of one. I know of one government lab that has done that, but, that's the only one that I'm aware of that has validated the mito for casework,
[00:28:37] Angela: And I'm gonna point out really quick too, that we are not advocating any particular type of platform, 'cause there's a couple of different companies that make this instrumentation. Softgenetics does software, you're not the only software company out there. that does it. There's different tools and there's different software on the market. And that's part of, I think laboratories are like, I don't know which one do I buy? Which analysis software is the best one for me to use? and it's such a complex laboratory test. It's not only do you have to get it and develop it internally and validate it and get all your procedures all in line and understand how to do all the analysis on it. It's then I have to go and explain it in the courtroom. And, um,right now, it's like that's a little bit further down the road it seems.
[00:29:26] Teresa Snyder-Leiby: Yeah,
[00:29:26] Megan Foley: Yes. Yeah. And that actually is one of our questions of when do you think this will be? we'll actually be running? at least like the crime scene type of samples. I think we'll see it with missing persons. that type of casework. I think that'll happen a lot faster.
[00:29:38] Angela: and I think we have,seen it for that kind of casework. It's definitely helpful right now in the identification of human remains for sure. Because there've been, you can see a lot of, like the national center for missing persons, they've definitely made some, Good identifications through this type of technology. But using it on a blood stain at the crime scene and comparing that back to the reference sample of a, persons of interest, it's a bit more complicated, cause it's much more expensive just to do one sample, let alone the crime scene sample and the reference samples for your suspects and your victims and everything. and,the equipment itself. If I remember correctly, cause I've bought, the Verogen MiSeq FGX and, um, you, I mean, there's some negotiating room on prices, but you're still looking at easily a quarter million dollars to get one of these machines into your laboratory and and It's not something you can't just run like one or two samples at a time. You, you need to run, I would think at least 24 samples to even break even on cost just to run it. So it's ...
[00:30:50] Teresa Snyder-Leiby: yeah. the the advantage though, is that they can run Y-STRs the new, Y SDRs and the mito all in the same sample. So the cost benefit would be looking at, as Megan was saying earlier, multiplexing them, and with the standard methods, we need the CE for the autosomals, for the Y-STRs and then for the Mito, so many different reactions. Whereas with the MPS they can all be done at the same time on the same sample. So that can help. So it can help bring down the cost. There was another article. I have to look for it where they were saying thatwas coming down in cost because of the chemistries becoming available. So again, this technology is developing, and getting things to the point where they are cost efficient is definitely a goal.
[00:31:44] Angela: I remember seeing a chart, you mentioned the human genome project earlier. I remembered seeing a chart of what would the cost was for sequencing way back at the beginning of that project? And it's $10,000 a sample or something ridiculous. and then,over time that it's just like a downward ski slope, because it's just, it goes right down and, like you can do a whole genome for a thousand dollars or something now. So depending on what the genome is, but yeah, you're right. So each individual you're gonna do your blood stain, if you, or your semen stain in a sexual assault case, The beginning part of it is the same. You still have to purify your sample. You still need to measure how much of your sample that you have, then you decide on what kind of technology you wanna use. and that's probably going to be something that labs will have to have a plan for to understand. Okay. So am I going to do my traditional, STR analysis on like Teresa mentioned the capillary electrophoresis system? Or am I, do I only have enough to do Y STR or if I have this new technology available to me, can I do STRs, Y-STRs, and mito all on the same thing? and it is definitely a cost benefit analysis, but it's not something that small laboratories would be able to do. I, you need to be a larger laboratory cuz you've gotta have that economy of scale I would think to be able to have the purchasing power, to get the supplies in the beginning. and it, these machines don't like to sit around doing nothing
[00:33:10] Teresa Snyder-Leiby: That's a good point.
[00:33:11] Megan Foley: they get crabby.they like to be busy. So there's allthese things that laboratories are struggling with. And I think maybe to relate this back to something for people who aren't actually in the laboratory, all of this technology is used very heavily in the biogeographical ancestry side of things. Now there are definite forensic applications for that, but people who are sending their DNA spitting into a tube and sending their DNA through the mail, they are benefiting from these technologies. Right?
Yep. Same instruments
[00:33:40] Angela: Yeah, it's the same instruments, the same technology. they're using some fancy softwareto do their analyses. And that's how they're tell, able to tell you that you're this percent african and or Sub-Saharan African and this percent, European and this percent, indigenous people. So that's where that comes from. It's this technology that's allowing that sort of thing to happen. so I wanted to switch more and let's talk a little bit. we talked around the technology and what we're doing with it, and where we're at. So let's say we get up and running with this technology in a laboratory and we've generated all of this data. What does Softgenetics do that can help us understand what we're looking at?
[00:34:20] Teresa Snyder-Leiby: Sure. well, with the MPS data you need software to import it because we're literally looking at terabytes, which is a term that makes my head just kind of implode, terabytes worth of sequence information. the software is based on a user interface where they bring in the data file and they have different settings depending on what their chemistry was. And if it's mito it will align all these sequences to a reference sequence. So there's a standard reference sequence that's used in forensics, and there are specific ways that it has always been lined up by this when analyzing Sanger data. This software enables the analyst to look at the whole depth of coverage. So they aren't looking at just a single sequence, as we were saying earlier, they're looking at hundreds or thousands of each of these little fragments lined up. And that way they can very easily visualize any minor mutations, which would put that person, in a different haplo group. that would be for the Mito. And then for the STRs, as Megan was saying, the capillary electropheresis or the older method they see more of a graph with, if there are eight copies at that one particular chromosome, it's a peak at that location. with the MPS software, gene marker HTS, then it aligns these and looks down through the depth. and so if half of those eights, maybe half, half came from mom and the other half, of course from dad, so their homozygous eight for their length. But if one of their parents had a minor difference in that repeat section, that's detected. And so it's presented then in a bar graph and also in a table that shows what's the balance. how much,is of this eight, is the same.And so that's where you've get this additional information is in looking, they call them ISO alleles because they have the same length. If I go back to, I love, Rudins analogy to the train you back to that the train is the same length so that they have the same number of those repeats but if half of them are, have a different, repeat sequence in them, it shows up and you expect to see that in a balance, pretty much half and half. and so itit's has the ability then when you're looking at mixtures to be able to look at at, if your minor contributor maybe has a mutation in it. It would show up as a separate, eight allele. so it's going to make it possible to tease out more information from mixtures.
[00:37:03] Angela: I have to say, I think that's one of the things that got me most excited when I first started learning about MPS way back was the ability for mixture deconvolution, the ability to discriminate between individuals, 'cause if you have a lot of times in sexual assault cases, you've got biological relatives that are in the same mixture. And if you've got these ISO alleles in there that will help you be able to the tell close relatives apart. And there was no way for us to do that with the traditional STR testing at all. This is like,the upholstery in mom's train car had the blue seats, but the, their daughter has green seats or something. That's like we're talking about with the ISO alleles, right? So the number of repeats are the same. It's just the stuff that's on the interior is slightly different. And it's those slight differences that this technology is letting us see.
[00:37:52] Teresa Snyder-Leiby: Right, And So in the userinterface, the bar graphs are nice because they're very similar to the appearance of the previous methods, they call 'em electropherograms those graphs we were talking about with the peaks.Well, bar graph is very similar. so that's a nice visual, But then the table shows you the actual balance. because it's just a little bit of a minor, if it's single source sample, and there's a little bit of a minor difference they might have a mutation or might be some noise, and that's where Megan's work on validation comes in. How much of, How much is noise?
[00:38:23] Angela: Looking at these bars and I remember they're, they tend to be color coded. So it's a nice visual representation. So you can see there's this minor thing that's there and that's maybe 10% of the mixture and someone else is 90% on that, that allele are there ways for us to incorporate that into the statistical analysis as well? Megan, is that something you're looking at too? You've got that percentage, the minor contributor and the major contributor there with the ISO alleles, since we had that information, it'd be nice. do we weight the statistic in some way to be able to help with a weight to the evidence that we find.
[00:39:03] Megan Foley: Yeah, absolutely. The probabilistic genotyping softwares, that I mentioned will actually go in and look at the profile and, look at a bunch of different factors about a DNA profile, way more advanced than I, would wanna get into, but it can look at patterns that are common to see, and look at the percentages of people or percentages of the, peaks that are there or those sequences, and try to tease out different individuals and do what's called deconvolution. So that's a lot of research that we still need to be able to get the sequence data into the software. And right now, our basic, using, our regular STR with the CE type of, methods, we can do that because we're just, looking at that number, repeat that's there, looking at that percent intensity of the different contributors, but now do you think every piece of DNA we had before, there's a bunch of different things we have to consider when it's sequenced. so it's just, it's much, much more in detail, but we need to teach the probabilistic genotyping software, these new patterns, the new details that we can see in sequence data.
[00:40:15] Teresa Snyder-Leiby: yeah. The details, I mean, a real basic one is that by capillary electropheresis a person can have one allele like homozygous the same, or they can be heterozygous. but with the deep sequencing,that person that's heterozygous, if they have ISO alleles, they might have four alleles at one locus and still be a single source. so yeah, it's, as Megan said, a lot more information, that's just one little part of it that needs to be incorporated to be able to,to make use of all that information.
[00:40:46] Angela: So it happens quite often people will submit, cases to the laboratory and be like, it's a rush. I need it tomorrow
[00:40:54] Megan Foley: Oh, no.
that's not gonna happen with this technology.
[00:40:57] Teresa Snyder-Leiby: no. it's definitely developing, but it is moving forward at a very good pace. We've been working with MPS sequence here, with the research software, at soft genetics since 2007, I think, and my husband said, to me, he says, you gotta learn this next gen stuff. You're gonna apply that in forensic. I said, no forensic community has to move very methodically forward. I will be retired before, before they get there. and they've gone and made a liar out of me because it is definitely up and coming and it's moving.I think it's moving at a very good pace because it's not, it's very, just like forensic science. Everything It's very methodical because you're looking at something that is, is, legal application of science,
[00:41:41] Angela: Just general question... I think that this is especially true for public laboratories. They're overburdened, they've got more cases than they can handle. They've got long turnaround times and, implementing this tech type of technology. Isn't something that you can do overnight. It takes time. And I noticed that as laboratories started to adopt probabilistic genotyping as the statistical method that we use, try to use routinely for our standard STR casework that we do now, there've been some growing pains .With that where people have actually given incorrect testimony in court cases have gotten overturned partially related to mixture interpretation, differences in opinions between, the, state laboratories expert, and maybe an external expert. So basically our old technology with, or a routine technology with a new statistical method, we've got issues in the, in the field where people don't necessarily understand it as well as they need to in order to be properly applying it and explaining it in the courtroom. How do you see the MPS working out? Cause I, I have my concerns. Because we are exponentially more complex, not only in the laboratory methods a little bit more complicated, then you've got the data analysis, then we've got to present this data. What kind of, issues do you guys see coming from that? Maybe this is more for, for Megan since she's, more forensic lab person.
[00:43:19] Teresa Snyder-Leiby: Yeah, well, I was gonna say I was gonna say education, but yeah, Megan, in terms of just validation process and making transitions, that's your area.
[00:43:29] Megan Foley: Yeah. the validation I did when I first validated the system we were using and I was working in a research lab. It wasn't even a casework lab and I did my first run single source, repeatability, and I went, oh my gosh, there's a lot more we have to consider 'cause each,component of what we do in analysis, you have that much more data. so I think I, some of the growing pains I think are going to be similar based and I, based on the survey that we did too, and just, yeah, it's the education, it's getting everyone on the same page of understanding, now we have these ISO alleles, are we even ready to do statistics even on those yet, we're familiar with a basic allele. soI think the validation process is going to take longer and you're right. People of, some of our survey respondents too said, I'm still trying to validate prop gen, probabilistic genotyping. I can't even fathom bringing on this whole new instrument platform.luckily the hands on process it's. It can be automated, which is great. And it is, stuff we're already used to as benchworking scientists with, the types of pipetting and everything like that. but yeah, I do think, getting to court, it's gonna be the same hiccups, which, hopefully some of the research that's being out there hopefully will help smooth some of these out. so that, labs are going in it together instead of just trying to deal with it on by themselves, you know? Yeah. Yeah.
[00:44:55] Angela: So the education it's definitely something that, like going to this workshop, that's free at AFDAA would be very beneficial for people. And then also I would encourage anybody who's listening that's a legal professional, don't rely on the laboratory to keep you informed. You should take the time to try to go to these workshops and maybe talk to companies like Softgenetics and get a little bit more idea, researchers like yourself, Megan, that are trying to work on it, to get a better understanding of where the field's going. Especially if you're somebody that's on a cold case integrity unit, this type of information might be good for you to have, especially if you're going through old evidence, trying to see, is there something new available that I might be able to do? yeah, 'cause I think it's funny, I'm remembering at the beginning of the podcast Teresa, you mentioned RFLP. the original DNA fingerprinting and I've been in court and had people, and maybe even in the past five years, and someone's, the prosecution starts trying to introduce DNA evidence in the courtroom and they're asking old questions from the RFLP days. So the technology's improving, like you said, forensics is slow and methodical, but at the same time.
[00:46:06] Teresa Snyder-Leiby: Yeah.
[00:46:06] Angela: we're still faster than our customers.
[00:46:09] Teresa Snyder-Leiby: And, and another point of, of that is a lot of what Sarah and I, and, the other support people here do is just like online assistance and training. And so we've worked with public defenders and project innocence, where they didn't do the wet lab production, but they have the data files. And so by putting them into the software, they can then visualize it also. So it's not limited. The software use is not limited to the laboratory, but to anyone who is needing to evaluate or Use the, the sequence information. that's really awesome. So that's, a resource for people that non-lab, people go to you and get some help.
Yes. Yep. Yep. So we can show them how to use the software. We currently don't do analysis as a service, but we do education so that if they want to be users of the software they can learn how
[00:47:02] Angela: that's really amazing that you guys are able to help educate that way. we are at an hour already on our recording, isn't it fun?
So
[00:47:11] Megan Foley: Is that fun?
[00:47:12] Teresa Snyder-Leiby: yeah.
[00:47:12] Megan Foley: Nerdy fun.
[00:47:14] Angela: yeah. So did anybody have any closing remarks that they wanted to make? Sarah is shaking her head.
[00:47:22] Teresa Snyder-Leiby: Sarah. And I are like counterparts where they can't keep me quiet and we have to encourage herto, speak. 'Cause you havethis experience, Sarah.
[00:47:31] Angela: One on one help phone a friend.
[00:47:33] Teresa Snyder-Leiby: Yeah, there you go. Exactly, Exactly. And patience unlimited. That's that's another one of her strength
That that's a good virtue to have. Absolutely. Having the workshop open to everybody. we're gonna try to get a variety of different speakers. so some from universities, one from a lab in New Zealand just trying to give more of an overall picture. So becausethe, technology is coming down the line at a nice pace and it's something that is an, a resource, people for their continuing education. I mean, as they get it into the universities, people now at universities are trained in PCR and CE so this is gonna be another layer added to that, but there's probably at least a 10 year gap between, when it's part of the normal education, formal education process. So workshops are an excellent way resources from, like you said, the companies that make the software, are resources for helping companies that make the chemistries the instruments are another resource.
[00:48:43] Angela: it's really good that you're bringing in, speakers from overseas as well, because it's. it's a global thing that we're looking at this technology and, like Megan said earlier that, EV everyone's, we're all in the same boat here, we're all trying to figure out the same thing and we're working together for that. And let's all learn from each other, like someone else. I, oh, I didn't work this way for me. So I had to try it this other way. and Megan, you're gonna talk more about those survey results from your research projects?
[00:49:14] Megan Foley: Yep. Yep. And some, so some solutions I at least know of right now as well. Try to give you those too. Yeah.
Yeah
[00:49:21] Angela: And remember, if you're not not part of the solution, you're part of the precipitate,
[00:49:25] Teresa Snyder-Leiby: we are having an nerd conversation. You gotta love it. gotta love it.
[00:49:29] Angela: Okay. And on that nerdy note, thank you ladies very much your time today. I appreciate it. And if anybody who didn't get that joke, go look up the word, precipitate and solution in it will make sense for you, but it's not funny anymore, if you have to explain it. alright, so thank you very much, ladies. I'm gonna get this edited and up as soon as I can. I hope to have this up before the workshop goes, but if anybody who's interested in attending the workshop. You can visit AFDAA.org and click on meetings and workshops. And that will take you to information about the main meeting, the workshop and links to register, but you'll need to register very quickly because it's coming up very fast. And if you wanna make sure you have a cookie and a drink, then you need to R SVP for the meeting. with that, thank you ladies. And, I hope everyone has a great day.
[00:50:22] Sarah Copeland: Thank you.